Metabolic Pathway Design Technology
Tomokazu SHIRAI
Institute of Physical and Chemical Research (RIKEN)


Tomokazu SHIRAI
Institute of Physical and Chemical Research (RIKEN)


We aim to propose the design of microorganisms that produce useful compounds with high efficiency, by metabolism design using a metabolic reaction model based on genomic information (Genome Scale Model: GSM). We are working on the practical application of a highly accurate prediction tool into which omics data are introduced, by linking individual metabolic pathway design tools.
For the microbial production of useful compounds, it is essential to have a technology that optimally designs "metabolism" including not only the intracellular carbon flow but also the balance between energy production and consumption and the balance between oxidation and reduction. This is because such a technology allows the understanding of the intracellular phenotype and the information obtained can be applied to the metabolism design of the target cell and to subsequent breeding. However, there is a limit to the human brain's capacity to think about more than 1,000 metabolic reactions occurring in a cell. Therefore, the computing power of computers is indispensable. Especially in recent years, due to the acceleration of the annotation process by the innovations in the genome sequencing technology and information processing technology, it has become possible to describe all metabolic reactions on a computer at the genome scale level. By this improvement, a technology for predicting the metabolic behavior of microbial cells in a certain environment has been established (genome scale model: GSM). Currently, research is being actively conducted by systematically performing the processes from cell metabolism design using GSMs to verification by actual experiments, in order to improve productivity to produce the target compounds in a high-throughput manner. In this project, we have developed a system that automatically prepares metabolic reactions from genomic information. However, the existing GSMs cannot predict or design biosynthetic pathways for non-natural compounds. In addition, design using metabolic reactions that occur in non-host cells is difficult.
In this project, we have developed metabolism design tools that can serve as a technology to solve these problems, and are applying this technology to useful microorganisms.
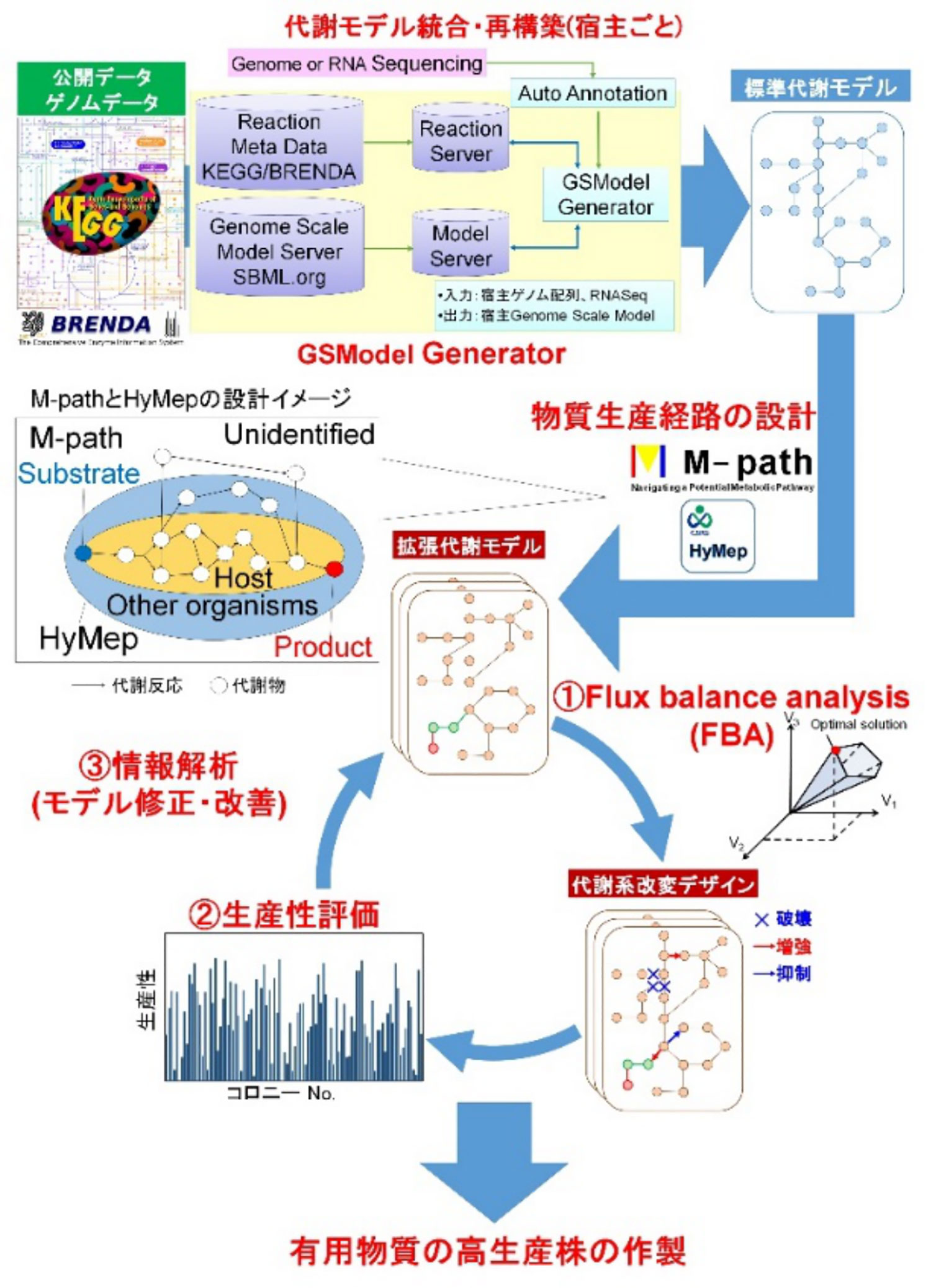
We aim to develop a highly flexible metabolism design system that meets the needs of the industry and research institutions, by operating a metabolism design system that functions interactively with each metabolism design tool developed.
We aim to provide useful information on metabolism design toward the production of target compounds.
Last updated:December 25, 2023