Protein Thermostabilization by MD Simulations
Tomoshi KAMEDA
National Institute of Advanced Industrial Science and Technology (AIST)


Tomoshi KAMEDA
National Institute of Advanced Industrial Science and Technology (AIST)


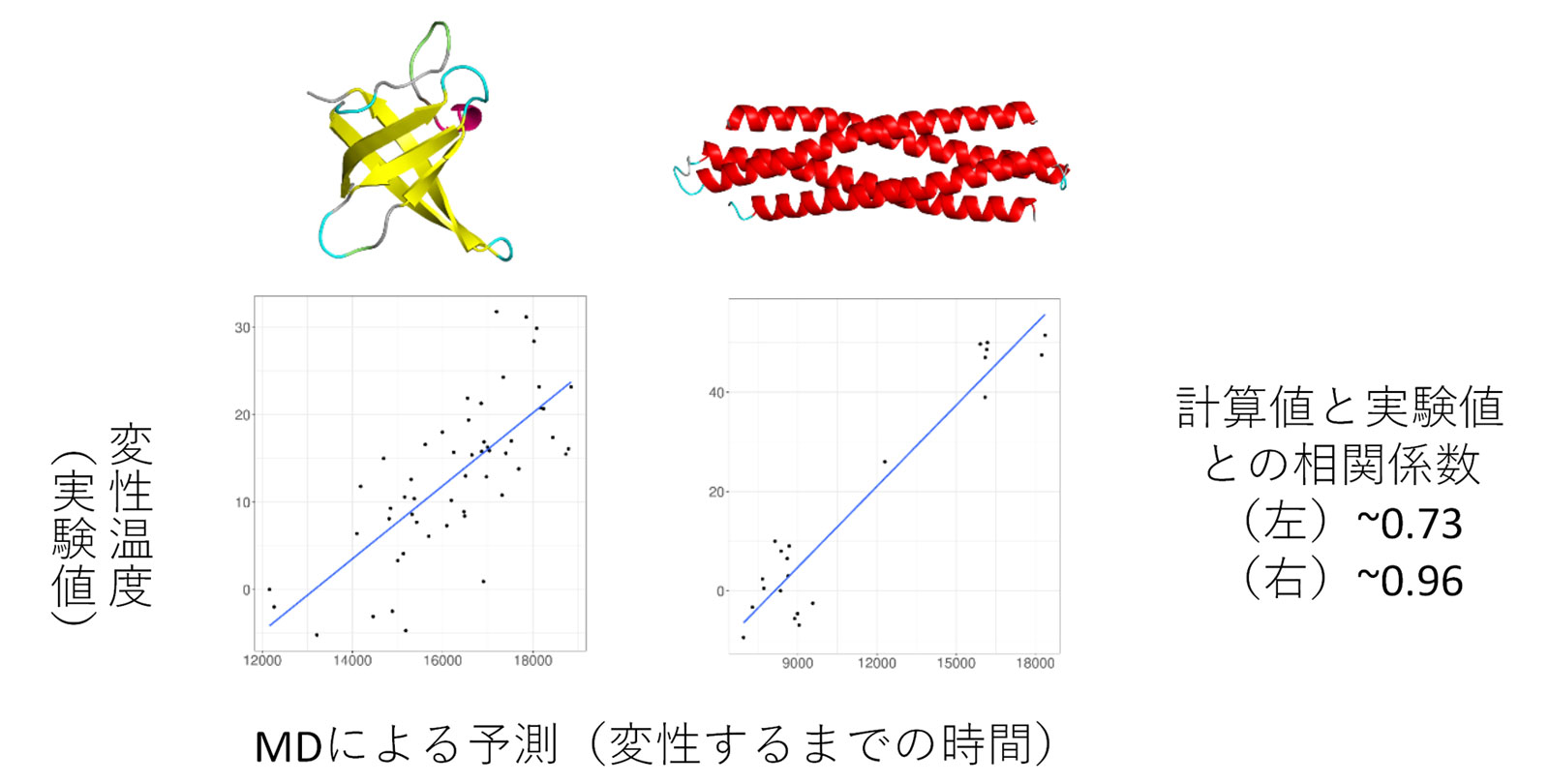
We discovered that the thermal stability (degeneration temperature) of protein variants can be predicted with high accuracy by analyses using molecular dynamics (MD) simulations. We designed thermally-stable variants based on this method and verified an actual increase in stability.
In this research, we worked on protein thermostabilization (increase in thermal stability) using MD simulations.
Protein variants were simulated at high temperature, and the time to degeneration was estimated.
It was found that the estimated time correlated well with the degeneration temperature obtained experimentally.
Therefore, various variants were simulated to predict the variants that are likely to increase thermal stability.
Actual increase in the thermal stability of these variants was confirmed by calorimetric experiments.
This method can be applied to any enzyme whose three-dimensional conformation has been determined.
Last updated:December 25, 2023