High-throughput (HTP) Transcriptome Analysis Technology
Yasuo MITANI
National Institute of Advanced Industrial Science and Technology (AIST)


Yasuo MITANI
National Institute of Advanced Industrial Science and Technology (AIST)


In order to establish a highly reliable transcriptome analysis technology, we intend to develop various spike-in control materials, and to perform verification within the project first. Furthermore, we are developing various technologies aiming to apply them to industrial microorganisms.
In order to establish a highly reliable transcriptome analysis technology that is applicable to various industrial microorganisms, we will develop nucleic acid reference standards for spike-in control and verify their utilization strategies. In addition, for that purpose, we will establish a simple method for the valuation of the nucleic acid reference standards.
In some industrial microorganisms, the production level of a targeted useful material increases especially at the end of the culture period. However, it is difficult to extract RNA in good condition from such samples. Therefore, we are developing a technology that allows the transcriptome analysis, which has been difficult to perform so far, by establishing a technology for selectively analyzing only non-degraded RNAs.

スパイクイン用核酸標準物質の開発と検証
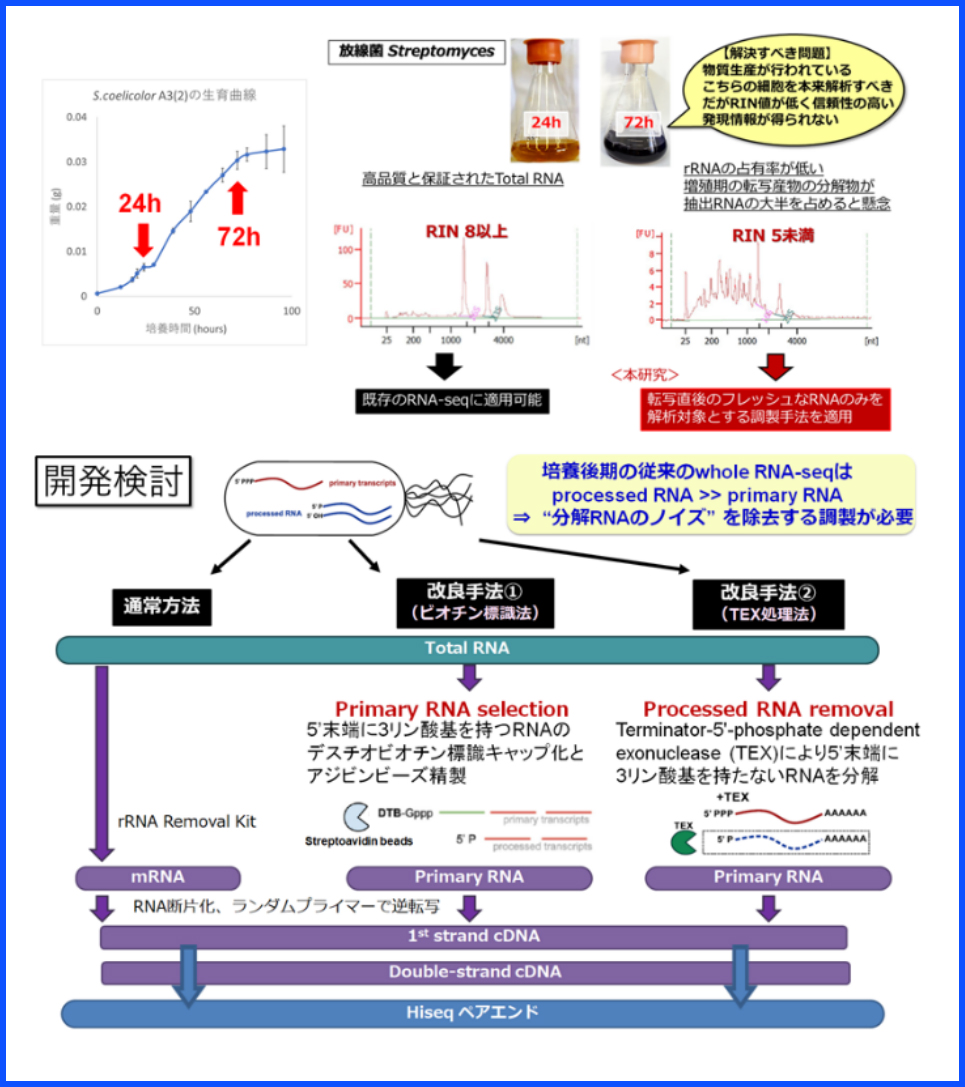
未分解RNA選択的調整技術の適用
Today, to improve the efficiency of material production using microorganisms, RNA-Seq transcriptome analysis is an essential technology element. Therefore, while various kits are available, it is necessary for each user to confirm the applicability of these kits, and many users tend to determine the applicability just based on their experience. In addition, also in terms of obtained data, the results may become meaningless unless a proper quantitative comparison between samples is feasible. We are developing technologies so as to make available the technologies and know-how that can serve for solving issues.
1)Mitani Y. et al.: Smart Cell Industry, CMC Publishing, (2018)
2)Noda, N. et al., Anal. Chem, 90, 10865-10871 (2018)
Last updated:December 25, 2023