Sample Non-destructive Cell Evaluation Technology
Nobuhiko NOMURA
Professor
School of Life and Environmental Sciences,
University of Tsukuba


Nobuhiko NOMURA
Professor
School of Life and Environmental Sciences,
University of Tsukuba


It enables the non-destructive identification of cell types at the resolution of a single cell, using a cellular autofluorescence pattern as the index, and the estimation of the metabolic state of the cell.
"Cell evaluation technology" is a core technology used for cell breeding technologies, differentiation induction technologies from stem cells to target cells, and technologies for the synthesis of artificial cells using a synthetic biological approach. However, it requires a lot of time and labor. For example, in order to evaluate the phenotype of useful material-producing bacteria, the useful material-producing bacteria that have been bred need to be batch-cultured, and the useful materials produced by the bacteria are extracted and quantitated. Therefore, the complicated work associated with cell culture time and quantitation is the rate-limiting factor for breeding. In addition, the existing cell evaluation technologies include fluorescent protein expression, antibody staining, etc. The majority of these methods require a process that modifies or destructs cells. There are few methods that evaluate the properties of living cells in a non-destructive and comprehensive manner. Moreover, the existing cell evaluation methods evaluate the average phenotype of a cell population. Accordingly, the phenotype of cells with some different traits that appear in association with proliferation and differentiation is overlooked. In order to select the target cells only from a heterogeneous cell population, a method that can evaluate the phenotype of each cell is required.
Therefore, we have recently developed an innovative cell evaluation technology, the confocal reflection microscopy-assisted single-cell innate fluorescence (CRIF) method, which can identify cells "non-destructively" at the "single cell level" and estimate the cell's metabolic status, using a cellular autofluorescence pattern as the index.1
Intracellular proteins and metabolites emit autofluorescence of various wavelengths and intensities. The autofluorescence pattern, which is formed by the synthesis of respective autofluorescence, functions as a "fingerprint" that expresses the properties of each cell. In the CRIF method, information on cell position and morphology is obtained by reflection microscopy, and cellular autofluorescence information is obtained by confocal laser microscopy.2,3 Then, by performing image analysis for each cell, the CRIF systematically and comprehensively extracts autofluorescence information for each cell, and reconstructs the information as an autofluorescence pattern. Thus, a "cell's fingerprint" that identifies each cell can be obtained. Moreover, it has been clarified that the application of the "cells' fingerprints" to various types of machine learning enables the construction of a machine learning model that reflects each cell's underlying characteristics in the autofluorescence pattern, and also enables the high-accuracy identification of cell types and the prediction of the metabolic state (Fig. 1). Previous studies allowed the identification of cells at different growth stages and the discrimination of Pseudomonas aeruginosa and Escherichia coli cells with a single gene mutation (Fig. 2).
The CRIF method, which can identify "cells' fingerprints" having almost infinite variations, has a potential to identify dozens (even hundreds, in principle) of cell types and metabolic status. In addition, since this is a simple method that can analyze the properties of intact living cells at the single cell level, it can be the key technology that improves the efficiency of the cell property evaluation, which is essential for cell breeding technologies, stem cell differentiation induction technologies, and technologies for the synthesis of artificial cells in various fields. At present, we are developing a technology for efficiently separating target cells after evaluating the properties of cells by the CRIF method, aiming at practical application of the CRIF method in screening. In addition, in cooperation with a Japanese microscope manufacturer, we are also developing a high throughput screening system that can perform a series of processes in the CRIF method (i.e., microscopic observation, extraction of cells' fingerprints, and evaluation of cell properties by machine learning). We aim to establish the CRIF method as a highly versatile cell evaluation technology so that it can be utilized in a wide variety of fields from basic research on, for example, microbial, plant and animal cell breeding, to applied research on, for example, regenerative medicine, in the future.
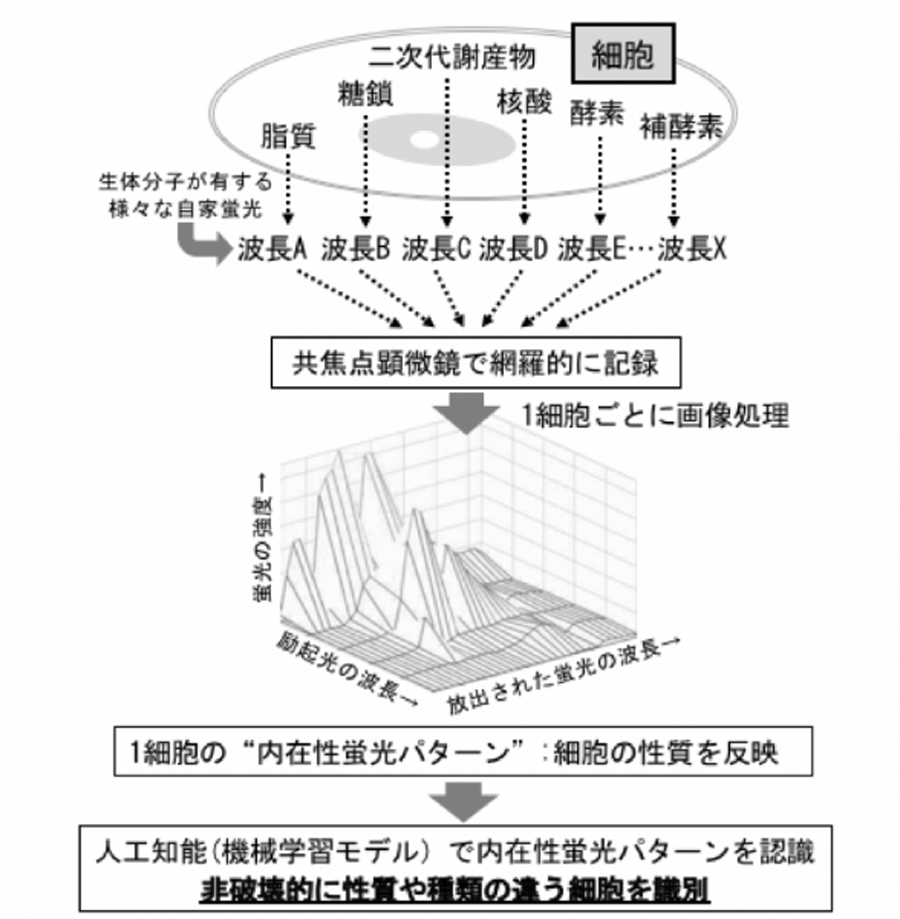
図1.CRIF(Confocal Reflection microscopy-assisted single-cell Innate Fluorescence)法の概念図
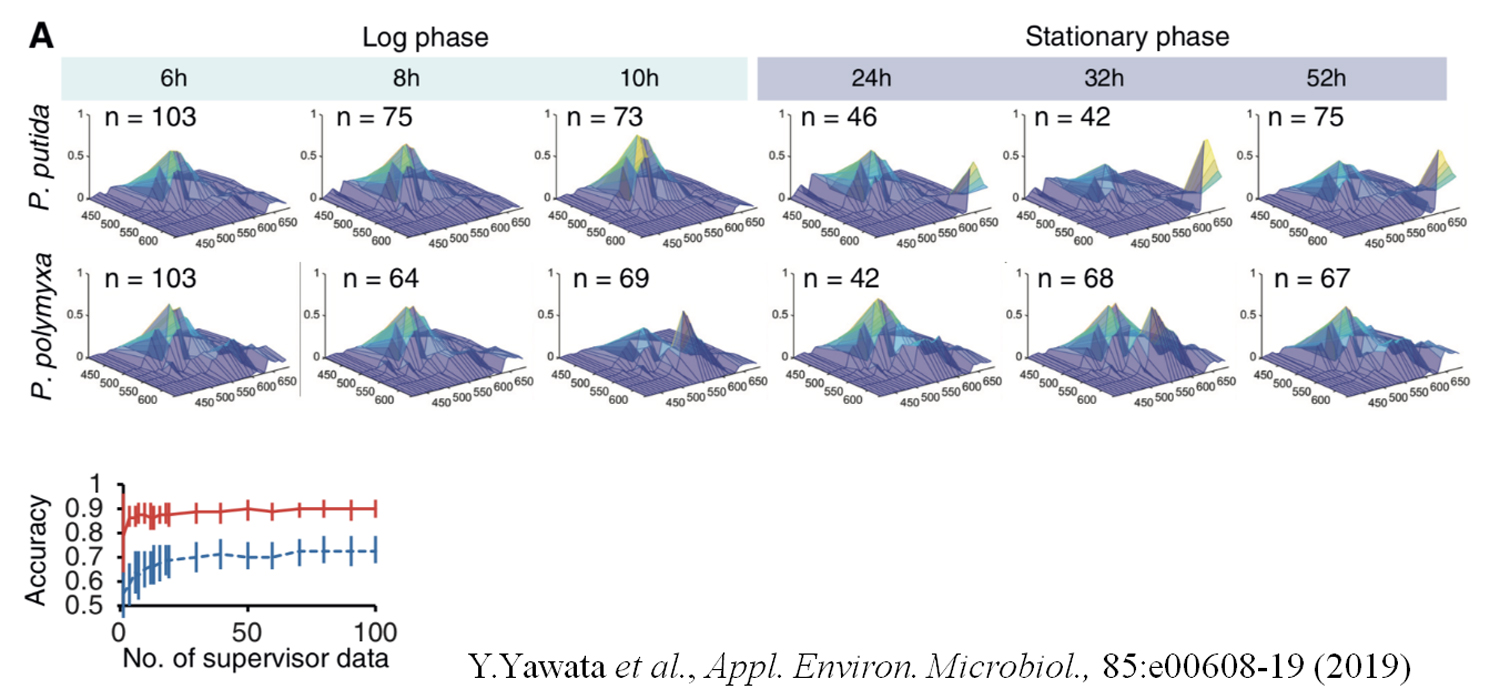
図2.P.putidaおよびP.polymyxaの各生育段階における平均自家蛍光パターン(上段)、ニューラルネットワークによる対数増殖期と定常期の細胞の分類(下段 赤:P.putida、青:P.polymyxa)
Japanese Patent No. 6422616 "Data creation method and data usage method"
Last updated:December 25, 2023