Enzyme Modification Design Technology
Tomoshi KAMEDA
Senior Researcher
Artificial Intelligence Research Center,
National Institute of Advanced Industrial Science and Technology (AIST)


Tomoshi KAMEDA
Senior Researcher
Artificial Intelligence Research Center,
National Institute of Advanced Industrial Science and Technology (AIST)


It enables proteins to have high functionality, specifically, a highly enhanced enzymatic activity, by modifying genetic sequences through a molecular dynamics simulation-based analysis.
The Smartcell Project aims to construct "smart cells" that maximize the material production capability of cells, and to create useful materials that are difficult to produce by conventional synthesis methods, as well as to achieve cost reduction and energy saving in the production processes. We focused on enzymes, which are actually responsible for material production in cells, and developed a method for predicting the enzyme modification site that improves their functions. An enzyme binds properly to a substrate that is used as a raw material for the material production and forms an enzyme-substrate complex to produce the target product (main product). However, depending on the shapes of the enzyme and the substrate, the reaction efficiency may be significantly reduced, or the purity of the main product may be lowered due to the formation of unintended products (by-products). Therefore, we aimed to maximize the material production capacity of enzymes, by making some enzymatic modifications to modify their structures to suit the main product formation.
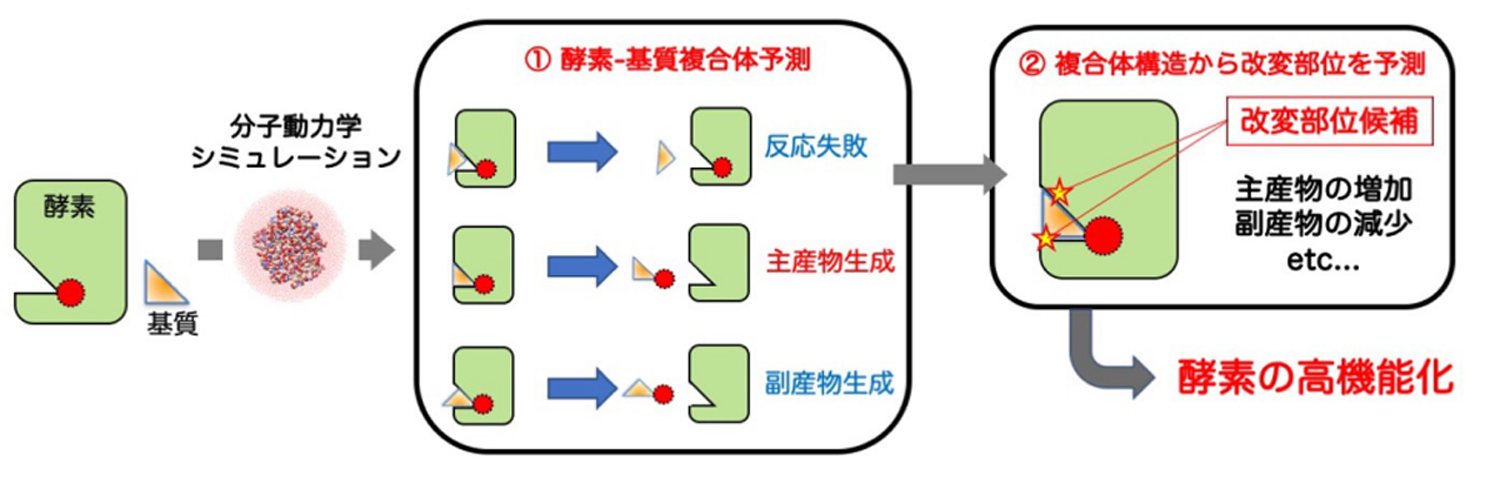
図1.酵素改変部位予測法の2つのステップ
This method for predicting enzyme modification sites can achieve high enzyme functionality by 2 steps (Fig. 1). In the first step, the adaptive lambda square dynamics (ALSD) method, which is a molecular dynamics (MD) simulation method developed by us,1,2 is used to exhaustively search for all possible enzyme-substrate complex structures at the atomic level. In the second step, from the information obtained by the ALSD method on the atomic level-structures of the complexes, the patterns of enzyme-substrate interactions are analyzed in detail, and then potential enzyme modification sites where the amount of the main product can be increased or the amounts of by-products can be decreased are proposed.
A major breakthrough in achieving high enzyme functionality was the use of the ALSD method,1,2 which is a powerful simulation method. In the MD, the structures of enzyme-substrate complexes are searched, by reproducing the environment of the enzymes, substrates and the solvents surrounding them, such as water molecules and ions, in the computer in order to track the time-series changes similar to reality. However, the conventional MD method requires a lot of computation time and computational resources to search the complex structures, and therefore it was very difficult to apply the method to actual enzyme modification research. On the other hand, the ALSD method is a modified MD method that can significantly promote structural changes at specific sites (such as substrate binding region) arbitrarily selected. In the currently ongoing research, various complex structures occurring during the formation of the main product or by-products are exhaustively searched, by promoting structural changes of the substrates inside the enzyme's pocket, and the patterns of enzyme-substrate interactions are analyzed in detail. As a result, it has become possible to predict the enzyme modification site at a simulation time of several days to about 1 week.
An actual application example is presented in Fig. 2. Although this enzyme has high reaction efficiency, it produces numerous by-products, resulting in low yield of the target compound, which accounts for only 13% of all the reaction products. By using this method, however, we succeeded in increasing the yield to about 70%. In addition, we succeeded in increasing the amount of the target compound produced up to about 6 times.
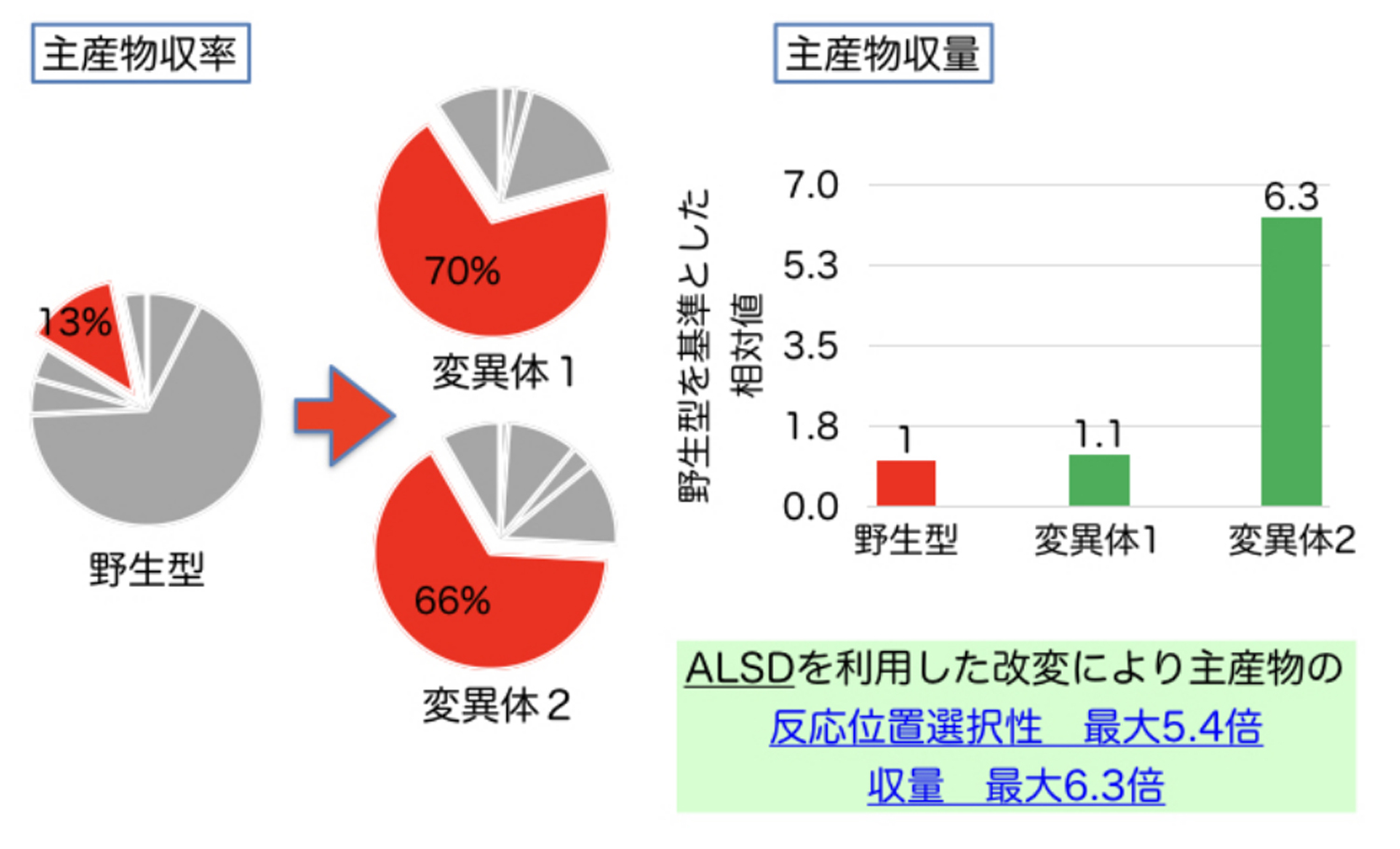
図2.酵素改変部位予測法によって提案された変異体の主産物収率、収量例
Last updated:December 25, 2023