High-accuracy Metabolomic Analysis Technology
Tomohisa HASUNUMA
Director and Professor
Engineering Biology Research Center,
Kobe University


Tomohisa HASUNUMA
Director and Professor
Engineering Biology Research Center,
Kobe University


It enables the identification of culture conditions and cells suitable for material production and also the identification of bottleneck reactions in metabolic biosynthetic pathways by highly reproducible multi-sample processing and comprehensive detection of water-soluble metabolites using robots. This clarifies the causal relationship between genetic modifications and the material production capability of cells.
Example of ApplicationUseful aromatic compoundsω-3 polyunsaturated fatty acid-containing lipids
Metabolomic analysis is a technology that can simultaneously identify the pool sizes of various metabolites present in cells. As the obtained metabolomic data reflect the growth environment and genetic background of cells, it becomes possible to identify culture conditions and cells suitable for material production by performing metabolomic analysis.1
In other words, measuring the isotope labeling ratio of metabolites over time after a stable isotope-labeled carbon (13C) or nitrogen (15N) source is added to the culture medium allows the comprehensive identification of metabolite turnover rates. The present method (dynamic metabolomic analysis) also allows identifying bottleneck reactions (rate-limiting reactions) in metabolic biosynthetic pathways, and if the rate-limiting reactions are identified, the cellular productivity can be improved by eliminating the rate-limiting reactions through genetic engineering.2
In recent microbial breeding, the following DBTL cycle is being established: (1) Designing the metabolism, etc. of recombinant microorganisms in a computer using biological information; (2) Building various types of microorganisms using standardized DNA parts and robotics; (3) Testing productivity to produce the target material in a short time, and (4) Learning the difference between the test outcome and the initial design computationally, which leads to the next design.1
When the metabolomic analysis is incorporated into the Test of the DBTL cycle, it can mimic a mechanism for improving productivity. Therefore, the quality of the Design is expected be improved and the number of cycles to be reduced (i.e., shortening of the period of breeding microorganisms).
However, the metabolomic analysis has problems in terms of throughput and data accuracy. More specifically, [1] the pretreatment process for extracting metabolites from cells involves complicated and time-consuming manual operations, with a low reproducibility; [2] the ion-pairing agent added to the metabolite detection system (LC-MS/MS) reduces the detection sensitivity and accuracy; and [3] since the number of metabolites that can be quantitated simultaneously is limited to about 100, important trace metabolites cannot be determined. The incorporation of metabolomic analysis into the DBTL cycle has not been achieved anywhere in the world.
Therefore, we first developed a robot that automatically performs pretreatment (Fig. 1). This automated pretreatment system consists of a sampling unit and an extraction unit. The sampling unit samples cell suspensions from a 12-chamber bioreactor at regular intervals, and then immediately stops the metabolic reaction. After collecting the cells, the extraction unit removes the medium and extracts metabolites.
When the performance of the extraction unit was evaluated using Saccharomyces cerevisiae as an experimental material, the process that took 3 hours for a skilled operator was shortened to about 1 hour. Since it can be operated overnight, it can achieve a processing speed (288 samples per day) that is 10 times faster than that achieved by humans.

図1.メタボローム解析用自動前処理ロボット
The automation has improved not only processing speed but also accuracy. The conventional pretreatment requires attention to many aspects, such as stopping the metabolic reaction, preventing leakage of metabolites immediately after the stoppage, removing the culture medium, efficiently extracting metabolites, removing lipid or peptide contaminants, and preventing degeneration of the extracted metabolites, and this has complicated the procedure and led to low reproducibility. However, our automated system has overcome these problems. In addition, the incorporation of a barcode reader has facilitated the management of the vast amounts of samples and data.
For the analysis after pretreatment, the S/N ratio has been improved by constructing an LC-MS/MS system that requires no ion-pairing agents. In addition, the method has enabled the isolation and detection of 158 water-soluble metabolites required for designing microbial smart cells.
In the metabolomic analysis, in which the identification and relative quantitation of various metabolites are performed for each sample, the amount of data processing is enormous. Therefore, we are constructing an information analysis system (Fig. 2) that assists in peak picking from chromatograms and projects the analysis results on a metabolic map.
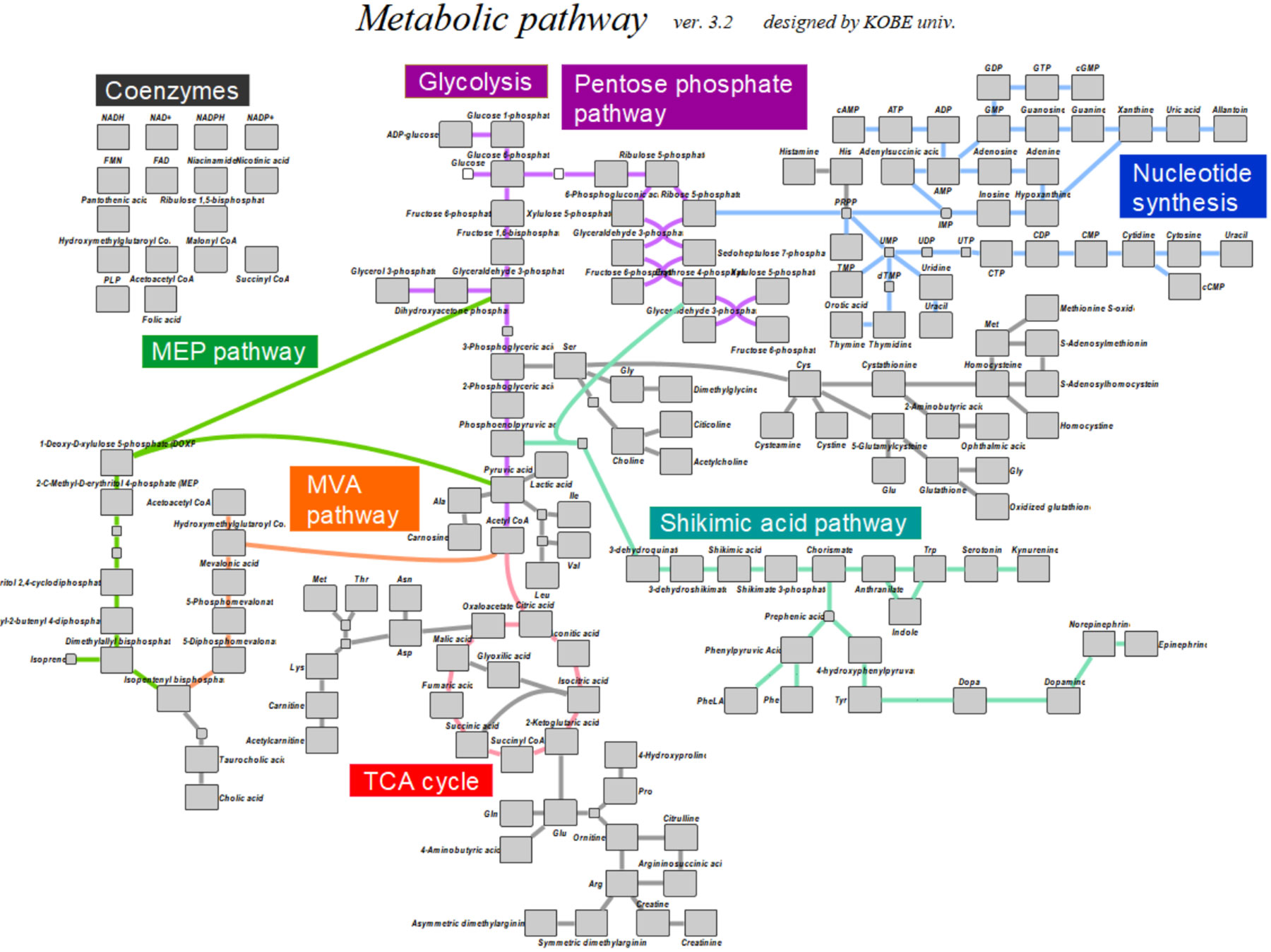
図2.開発したシステムにより測定可能な水溶性一次代謝物の代謝マップへの投影
Recently, with an increased interest in biotechnology-based material production processes, the evaluation of bioproduction has become more important. In microbial breeding so far, even though multiple metabolic modification strategies could be proposed, an optimal solution could not be derived due to a lack of adequate knowledge about the metabolic control mechanism.
We intend to elucidate the metabolic control mechanism, which has been a black box, by obtaining a large amount of highly reproducible metabolomic data through the development of this technology, and to develop a DBTL cycle that incorporates the mechanism analysis. The cycle is expected to create groundbreaking production strains (smart cells) successively in a short period of time.
Japanese Patent Application No. 2018-134171
Japanese Patent Application No. 2018-134174
Japanese Patent Application No. 2018-134169
Japanese Patent Application No. 2018-134177
Japanese Patent Application No. 2018-134179
Japanese Patent Application No. 2018-207920
Last updated:December 25, 2023