Quantitative Targeted Proteomic Analysis Technology
Fumio MATSUDA
Professor
Graduate School of Information Science and Technology,
Osaka University


Fumio MATSUDA
Professor
Graduate School of Information Science and Technology,
Osaka University


Highly sensitive simultaneous quantitation of the expression levels of dozens of proteins.
Example of ApplicationUseful aromatic compoundsω-3 polyunsaturated fatty acid-containing lipids
The Smartcell Project requires rational modifications of metabolic pathways in order to maximize the material production capability of cells. To modify metabolic pathways, the expression levels of enzyme proteins are increased or decreased by artificially modifying the genome of the host microorganism. Since the protein amount in cells involve various factors associated with transcription, translation and proteolysis, it is necessary to have a measurement method for rapidly evaluating whether the amount of enzyme protein has been adjusted as designed. Under these circumstances, we focused on the high-accuracy quantitative targeted proteomic analysis as an optimal method for the evaluation of smart cells. In this method, a trypsin-digested peptide mixture is separated by liquid chromatography and analyzed in the multiple reaction monitoring (MRM) mode of a triple quadrupole mass spectrometer. Currently, we are developing a targeted proteomic analysis system authentically made in Japan, in collaboration with Shimadzu Corporation (Fig. 1).
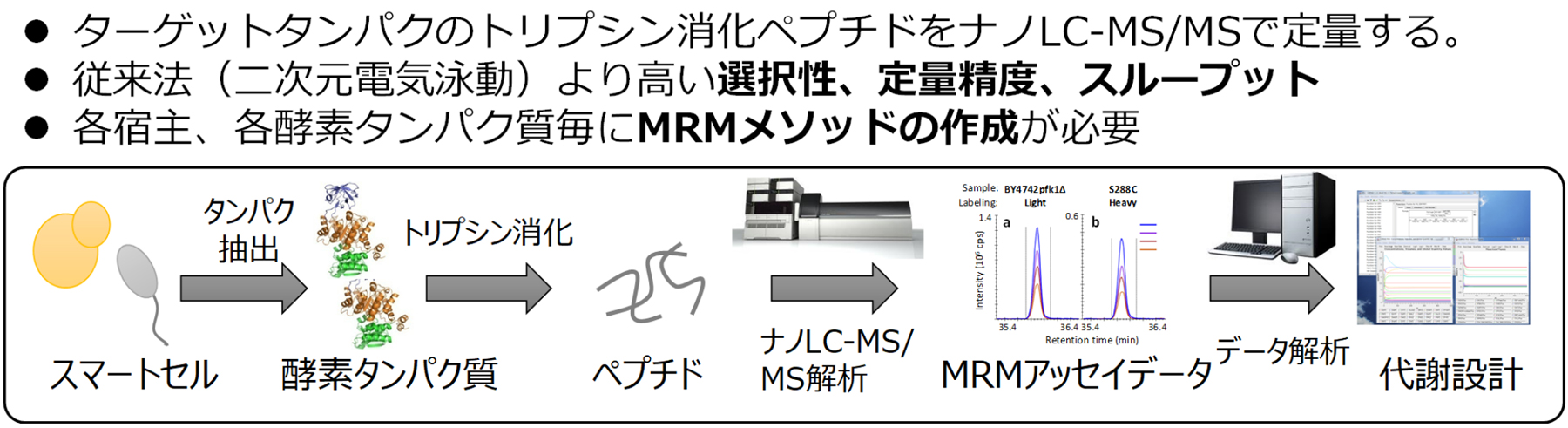
図1.定量プロテオミクス法の概要
The key points of the targeted proteomic analysis are establishing a sample pretreatment method and a quantitative analysis method (MRM assay method). First, for sample pretreatment, a crude protein extract of about 100 μL containing 50 μg of total proteins was used as a starting material. After a reductive alkylation of this extract, trypsin digestion is performed overnight, and the resulting trypsin-digested peptides are desalted by solid-phase extraction. This pretreatment method for Saccharomyces cerevisiae was effective against various useful microorganisms, such as oleaginous yeasts, Escherichia coli and Corynebacterium glutamicum.
Data were obtained by nano-LC-MS/MS (LCMSTM-8060, Shimadzu Corporation). This system, in which the number of MRM channels per second can be increased up to 500, has performance suitable for quantitative proteomic analysis. The current system, in which nano-LC is used for emphasizing sensitivity and the processing time is set to be 1.5 hours per analysis, has achieved a throughput of a sample size of 20 to 30, which is the size of a general analysis project, in about 2 days.
The multiple reaction monitoring (MRM) assay method selects 3 to 4 quantitative peptides that can be detected with high sensitivity by LC-MS from a large number of trypsin-digested peptides produced from the protein to be measured, and further selects 4 MRM sequences optimal for each peptide. Since the MRM assay method depends on the amino acid sequences of the protein, it must be prepared for each protein. To promote efficiency in establishing an MRM assay method, the number of reactions to be monitored needs to be reduced. Therefore, we have developed a new method to predict peptides suitable for quantitation from peptide sequences. This method, using a different approach from the conventional method, searches for hopeless peptides that are unlikely to be adopted in the MRM assay method. While minimizing the overlooking of suitable peptides, 30% to 40% of all peptides can be successfully removed as hopeless peptides.1 We will continue to promote the efficiency in constructing the MRM assay method, and this is expected to lead to the rapid construction of an MRM method and eventually to a rapid development of smart cells.
Using these methods, we have completed the construction of MRM assay methods for main metabolic enzyme proteins, such as E. coli, S. cerevisiae, C. glutamicum and oleaginous yeasts. The MRM assay methods we constructed are compatible with mass spectrometers of different manufacturers. Therefore, currently, we are building a database of MRM assay methods constructed in the Smartcell Project, so as to establish a platform that can be customized as required (Fig. 2).
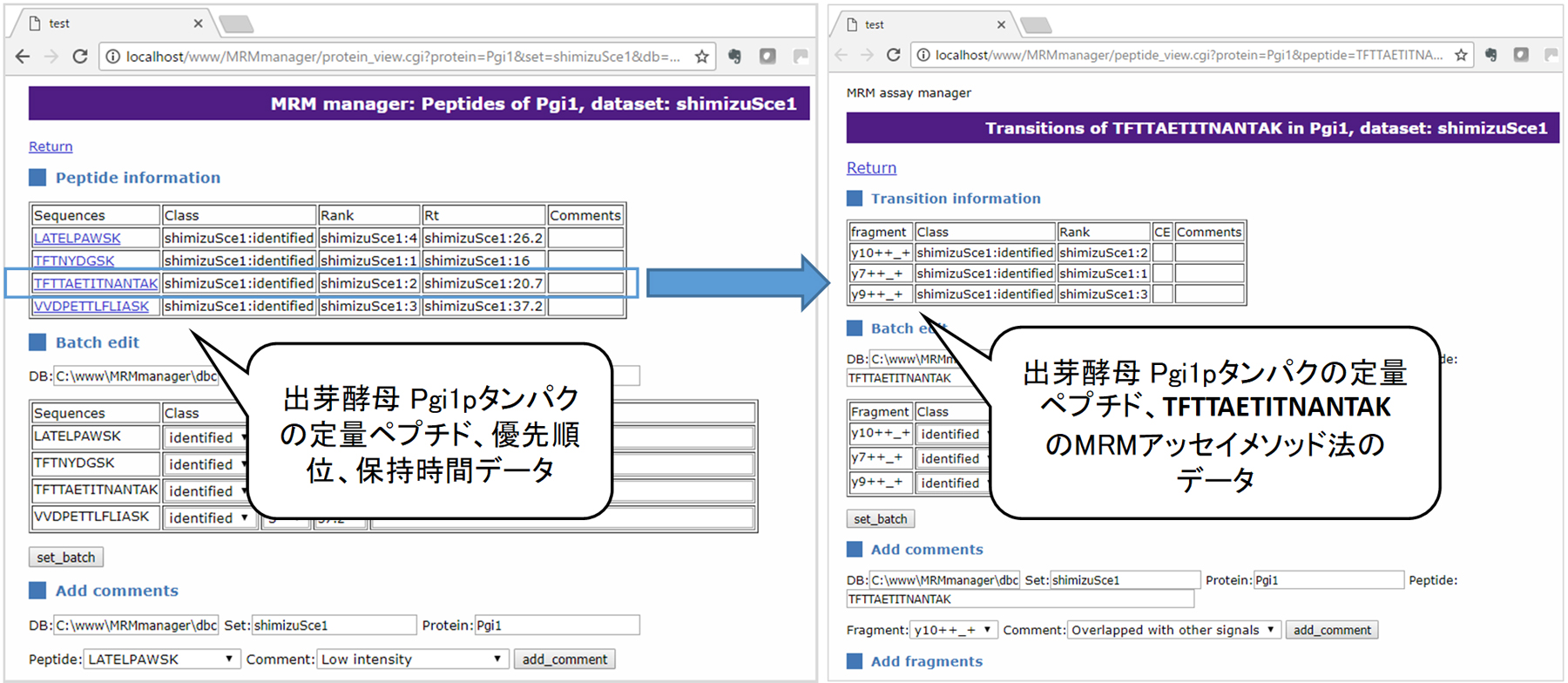
図2.MRMアッセイ法データベース
Last updated:December 25, 2023