Knowledge Extraction and Learning from Literature, etc.
Michihiro ARAKI
Program-Specific Professor
Graduate School of Medicine, Kyoto University


Michihiro ARAKI
Program-Specific Professor
Graduate School of Medicine, Kyoto University


This technology allows the extraction of useful knowledge for microbial development from literature and public data on smart cells, which enables the proposal of the next step to improve the current design, such as promising gene modifications or enzyme genes for increasing productivity.
Example of ApplicationUseful aromatic compoundsω-3 polyunsaturated fatty acid-containing lipids
The "Learn" process in the Design-Build-Test-Learn (DBTL) cycle of the smart cell development, i.e., the interpretation of various data/models, and the following process of creating the design hypothesis based on them, relies on the background of personal knowledge and on manual literature/database searches and surveys. Such a knowledge acquisition process dependent on individual skills is a rate limiting factor in the smart cell development, which interferes with systematic accumulation/discovery/reuse of knowledge, and poses a major issue to be resolved technically. For example, the metabolic pathway design, metabolism model construction and optimization, and search for enzyme genes and modification candidate genes largely depend on knowledge extraction from the existing literature and database information. In many cases, existing databases on metabolic pathways and enzyme reactions do not contain sufficient information necessary for smart cell development. Therefore, there was plenty of room for reconsideration regarding the extraction of literature knowledge for smart cell development. In addition, with the recent development of machine learning and artificial intelligence (AI) technologies, it has become possible to extract new knowledge and patterns from existing data, and such technologies are expected to become applicable. Under these circumstances, this project works on the development of a knowledge base from the literature that supports metabolism and enzyme design proposals and the development of a machine learning technology focusing on enzyme gene search.
(1) Knowledge base development
In order to organize the individually acquired knowledge on microbial design and systematize it in a reusable form, we developed a technology to organize and accumulate the design history of microbial strains and the genetic modification content of each strain associated with the design history, in terms of information on purpose, means, and rationale. The "visualization" of the accumulated knowledge in a tree form along the modification history allows an overview of the entire design data to date and utilization of the data to inspire new hypotheses. In addition, based on the accumulated and systematized design history, the system calls up the associated knowledge extraction technologies, and presents useful information that leads to design improvement.
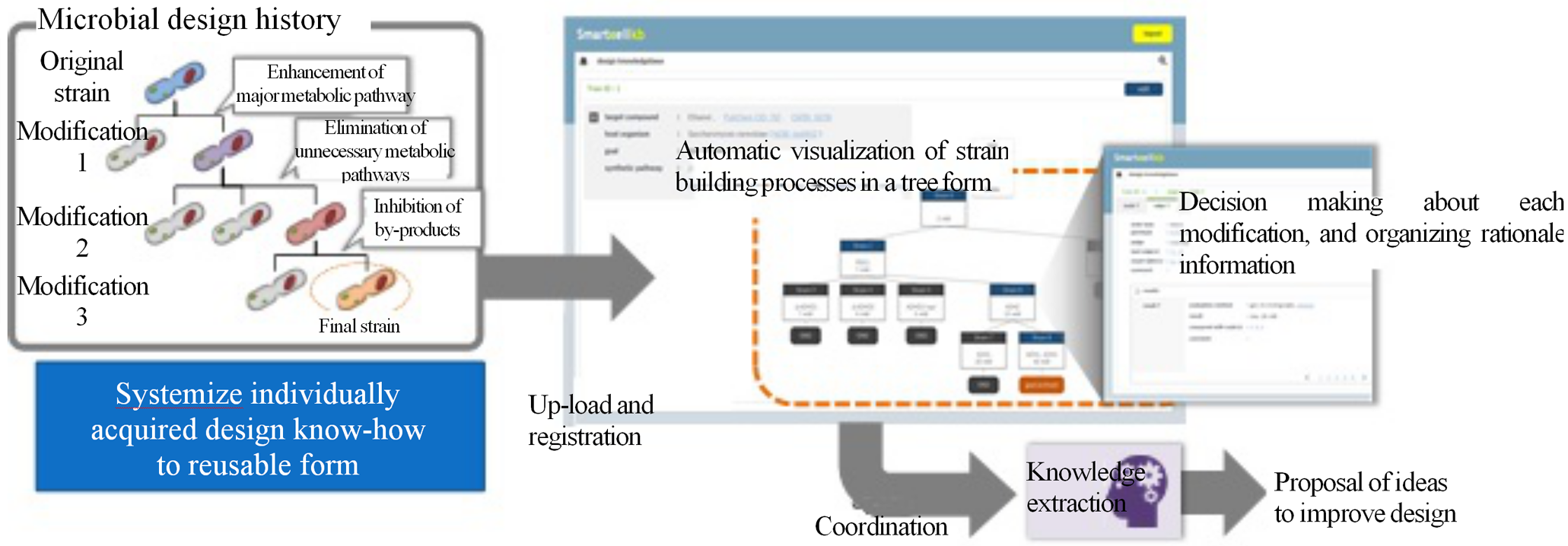
Figure 1 Knowledge base for supporting smart cell design
The knowledge extraction technology consists of smart cell literature automatic collection technology and promising gene recommendation technology. The former technology identifies characteristics of literature related to metabolism design and gene modification by natural language processing and literature search technology, and widely collects useful literature information for smart cell design. The latter technology extracts and proposes genetic modifications deeply related to the previous metabolism design/gene modifications from the collected literature information. These technologies allow the collection of literature information related to the design from existing strain designs accumulated in the knowledge base, and the proposal of gene modifications to be added to the current design. As a result, it can be expected to run the DBTL cycle more efficiently and reduce the number of processes required to create the desired smart cell.
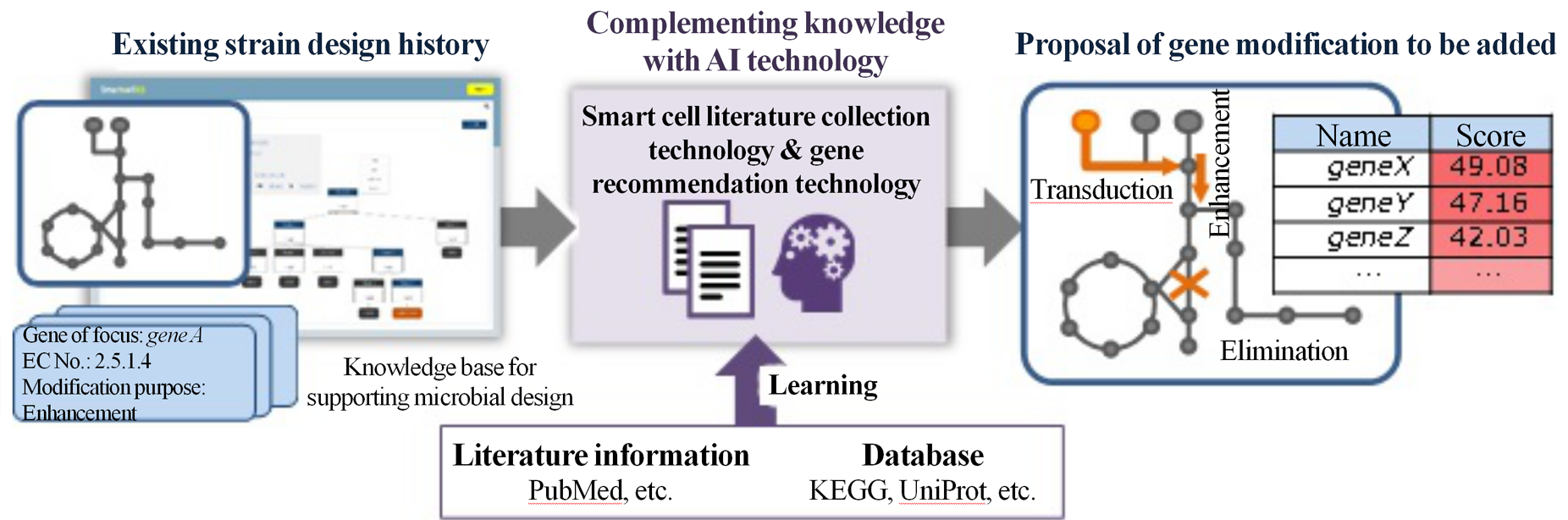
Figure 2 Gene modification proposals from literature information based on design history
(2) Enzymatic reaction data learning and activity estimation model
In each metabolic pathway created through a metabolism design process, multiple estimated enzyme gene candidates appear, regardless of whether they are unknown or known. Therefore, the selection of enzyme genes is an important challenge for actually constructing a metabolic pathway. This technology works as follows: First, it digitalizes the chemical structure and enzyme amino acid sequence of a substrate/product, and determines whether the enzyme regulates the reaction positively or negatively; then, it gives a score to test data having a new combination of a substrate/product and enzyme amino acid sequence, in order to determine whether it is positive or negative. Along with the conventional procedures for the machine learning and deep learning base, we also work on improving models by incorporating substantiation and verification data.
For example, we have developed a technology that provides guidance for enzyme gene selection by comparing the distribution of results judged by the machine learning method (Fig. 3). We also developed a technology2) for searching new metabolic pathways by predicting chemical structures possibly occurring after known and unknown enzymatic reactions, using a deep generative model that has learned the chemical structures of substrates/products (Fig. 4).
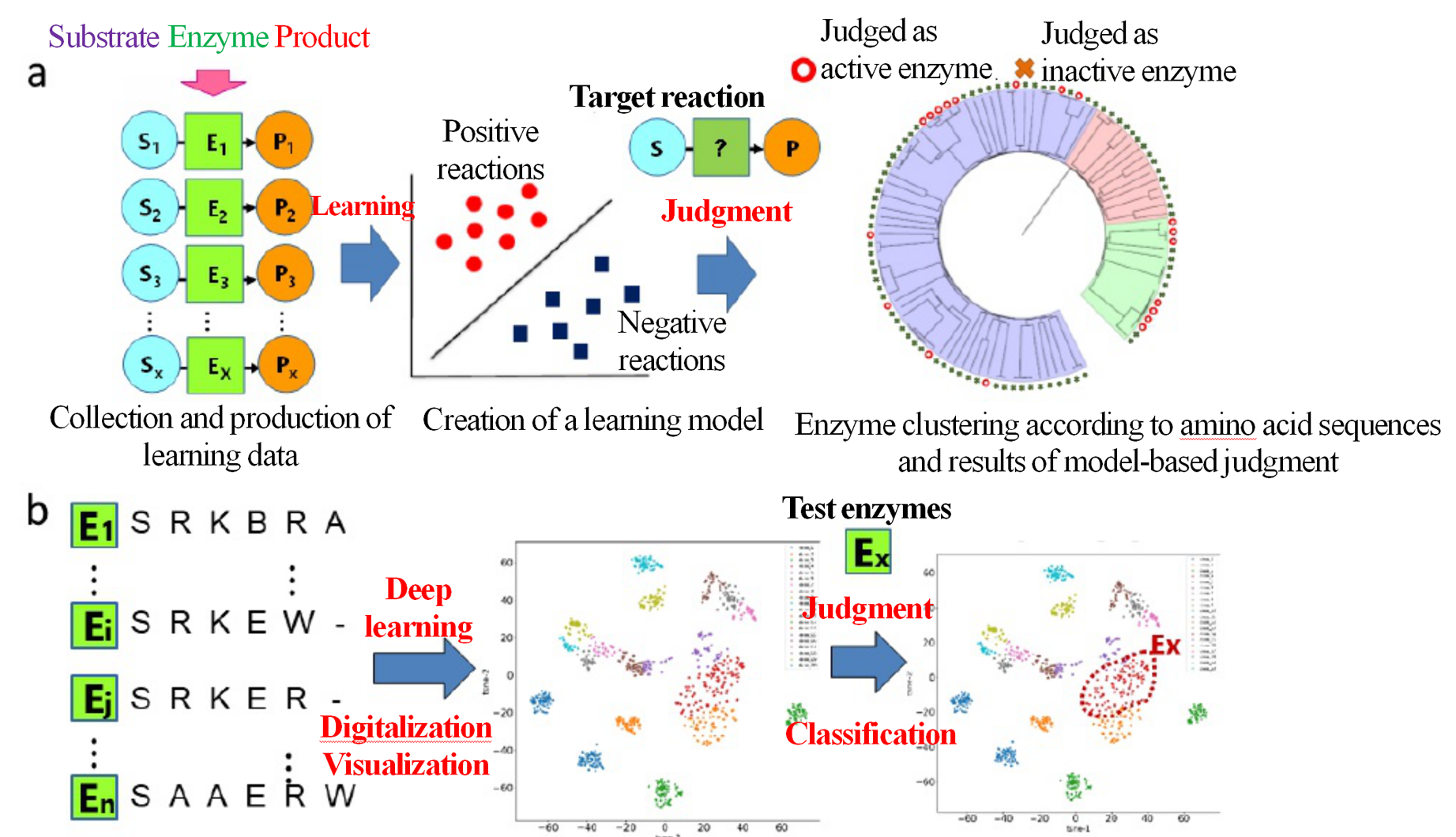
Figure 3 Learning based on enzyme reaction data and development of activity estimation model
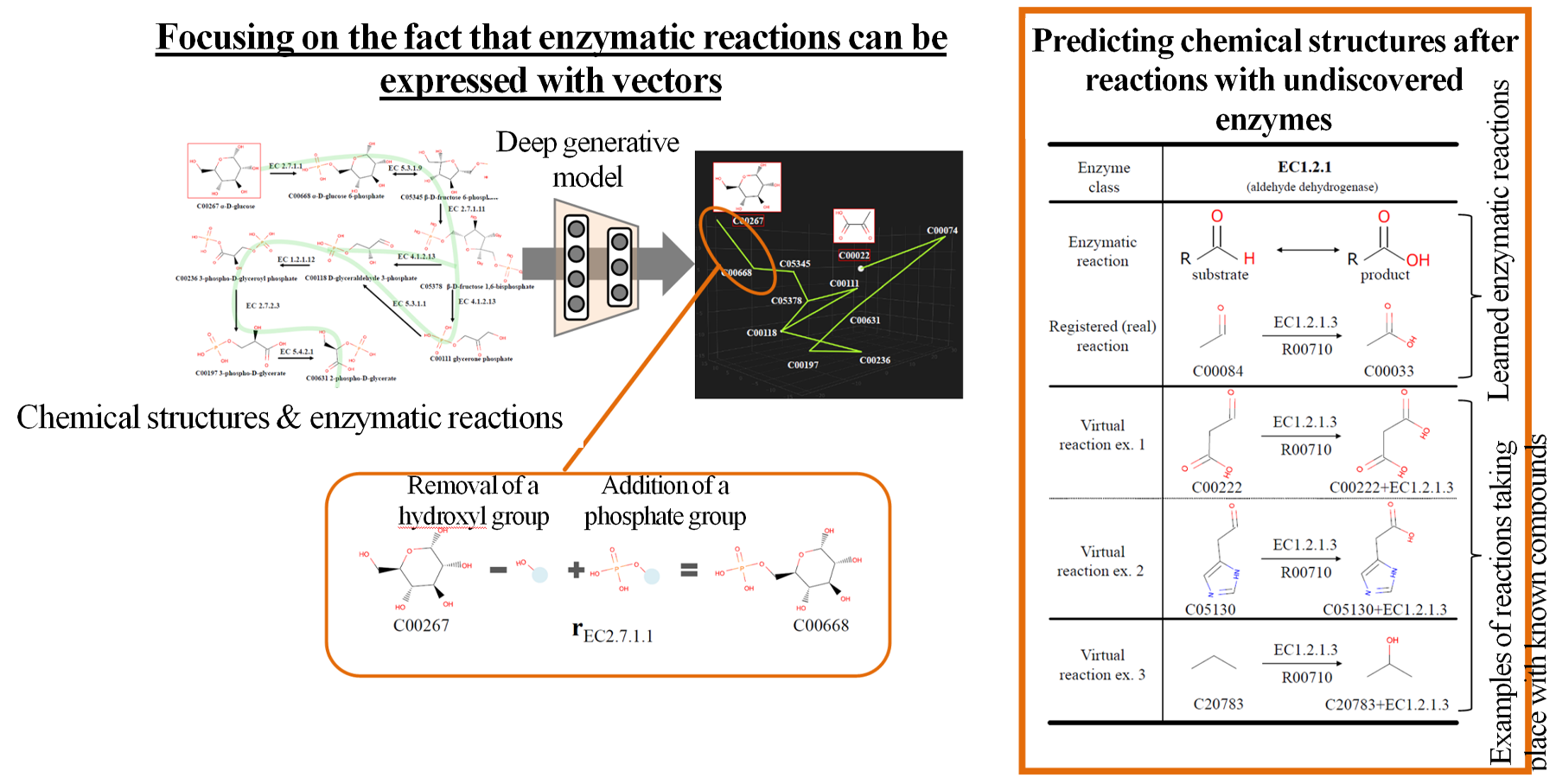
Figure 4 Search technology for new metabolic pathways using deep generative models
1) Watanabe, N., Murata, M., Ogawa, T., Vavricka, C.J., Kondo, A., Ogino, C., *Araki, M.: Exploration and Evaluation of Machine Learning-Based Models for Predicting Enzymatic Reactions, Journal of Chemical Information and Modeling, 60(3), 1833-1843 (2020)
2) Fuji, T., Nakazawa, S. and Ito, K.: Feasible-Metabolic-Pathway-Exploration Technique using Chemical Latent Space, Bioinformatics (2020)
Last updated:December 25, 2023